CCCM crystallographic coupled cluster method
What's about?
CCCM is a high-order CCM (coupled cluster method) code
for lattice spin systems at T=0 by Damian J. J. Farnell (DF).
Historically, this code is based on "foundations" laid by Ray Bishop
and John Parkinson (amongst others), and Chen Zeng's work in the 90's
on the high-order code. This code has been exploited and
pushed forward by Damian Farnell and Sven Krüger primarily. Indeed,
the current incarnation of the CCM code is still being used and
extended by Johannes Richter and his co-workers.
Because Damians time is being increasingly concentrated on other topics
Joerg Schulenburg (JS) did some improvements and necessary changes to the code
and set up this webpage for distribution of the most recent code.
If you have suggestions and code patches, send it to
Joerg together with a description what it does (better),
so he can include it in the official version.
News
- Jan16: 2400*2.4GHz and 20TB used (fix "HASH-Overflow happens"
by hash_bits=26 in collect.c) for 2D LSUB12, 4.2e6 clusters,
363e9 terms (RZ)
- Feb09: 4800 MIPS Cores on an SC5832 used,
S=1 Linear Chain SUB-14-14, 417min 720GB distributed memory,
Eg=-1.39374842, GAP=0.4214360
- Jun08: Scaling up to 1020 Itanium Cores on an Altix4700 (HLRB-II at LRZ),
Square LSUB12 1020*6h 640GB Eg=-0.668978086!
- Mar08: after CCM0511 a bug was introduced (missing terms33 for srcubo_spiral alpha!=0),
this is fixed now.
- Oct05: Joerg rewrote most of the code to make it better readable
and faster.
Speedup is up to 100 times now, download and enjoy ccm0509.
- Oct04: Joerg removed unnecessary disk usage and excessive output.
Opening this website.
Download
check signature using gpg --verify ccmXXXX.asc
- ccm1908.tgz v2019-08-13,
see ChangeLog (1.2MB, gcc8-fixes)
Signature)
- ccm1112.tgz v2011-12-09,
see ChangeLog (1.2MB, bugfix for angle search)
Signature)
- ccm1109.tgz v2011-09-06,
see ChangeLog (1.2MB, fix autoset of btLength v1002 Uc2++ LSUB7++)
Signature)
- ccm1107.tgz v2011-07-21,
see ChangeLog (1.2MB, 3 improvements for UseAlpha + EXIT_ON_ERROR)
Signature)
- ccm1104.tgz v2011-04-05,
see ChangeLog (1.2MB, 2*bugfix)
Signature)
- ccm1011.tgz v2010-11-27,
see ChangeLog (1.2MB, output file removed, see ChangeLog)
Signature)
- ccm1009.tgz v2010-09-17,
see ChangeLog (1.2MB, output file removed, see ChangeLog)
Signature)
- ccm1007.tgz v2010-07-19,
see ChangeLog (1.2MB, bugfix option --expects)
Signature)
- ccm1006.tgz v2010-06-22,
see ChangeLog (1.2MB, fidelity added)
Signature)
- ccm1005.tgz v2010-05-31,
see ChangeLog (1.2MB, more stable direct iteration for GAP)
Signature)
- ccm1002.tgz v2010-02-26,
see ChangeLog (1.2MB, now .c files, lot of simplifications)
Signature)
- ccm1001.tgz v2010-01-31,
see ChangeLog (1.2MB, new options and samples, code simplified,
Signature)
- ccm0911.tgz v2009-11-17,
see ChangeLog (Bug fix: spiral states + GAP)
- ccm0905.tgz v2009-05-26,
sample j1j2_square*.ccm corrected (striped state), see ChangeLog
(1.1MB, Signature)
- ccm0904.tgz v2009-04-24,
works better for triangular lasttice in a field, see ChangeLog
(1.1MB, Signature)
- ccm0901.tgz v2009-01-30
fix direct iteration (parallel) for excitations, see ChangeLog
(1MB, Signature)
- ccm0812.tgz v2008-12
add direct iteration (parallel) for excitations, see ChangeLog
- ccm0808.tgz v2008-08
Bug fix (for parameter alpha)
- ccm0807.tgz v2008-07-15
New Excitation Energies (some bugs fixed)
- ccm0806.tgz v2008-06-21
New Expectation Values and more (beta!, see ChangeLog)
- ccm0804.tgz v2008-04-08
(bugs, see ChangeLog)
- ccm0803.tgz
Bug fix for SrCuBO_spiral (CCM0605,CCM0607)
(840kB, 2008/03/31 Signature)
- ccm0607.tgz
abs(INT32_MIN)-Bug fix (2006/07/11 Signature)
- ccm0606.tgz
some improvements (2006/06/12)
- ccm0605.tgz
using memory (set ulimit -v to avoid swapping) (2006/05/31)
- ccm0511.tgz
experimental version, use with care! (2006/04/12)
- ccm0509.tgz
50-100 times faster (2005/10/05, len=794kB)
- ccm0410.tgz
less disk usage, less output (2004/10/21, len=964kB)
- Dissertation Sven E. Krüger 1999
(PDF-1.2, lang=german, 903kB, 101 pages)
ChangeLog
--- please also look at the ChangeLog file at the tgz package ---
31.05.10
- bounding=... in ccm-script removed (automatically computed)
- set GAP=0 if DI is not converged
- improve shift estimation for DI-GAP-code (EXPERIMENTAL=4)
every 2nd GAP was bad for J1-J2 models
- rewrite DI-GAP (power method, see wikipedia), making it more robust
- add option to set start vector (--startx=0..., see setStartX in solve.c)
sample: triangle.ccm sub-5-5 NR convergence
- better TermCompression (minimum size computed at runtime, bitshift used)
more compression by use of maxbits + bitshift, (cfg+term only once?)
ex: 5*18bit+4bit.term+Xbit.factor05=94bit+X=12Byte instead of 16,24,32B
- check number of different coefficients (for better compression)
switch on by TermCompression+=2 (smaller TERMSIZE, up to 50% less memory,
no precission loss, but save to disk may not work compressed! ToDo)
- print warning, when bounding triangle is set, bounding=... is obsolete now
- fix bug: LatticeSymmetries.number >= MAX_NUM_SYM
- add option --symmetries=... to set num symmetries (use carefully!)
- add option --expects=... to set max. num of computed expectation values
- fix: accuracy 1e-8 instead 1e-10 for j1j2_square_Neel J2=0.2 (float used)
- output Mag iterations if error grows (divergence)
- add options --subm and --subn, overrides script m and n (more flexibility)
- add option --mom (weight or momentum for direct iteration, macro removed)
- add options --increment --minimum --maximum for parameter range
to have the advantage to use the same script file for different batch jobs
- dont exit if bra-vector does not converge (useful for Delta scans)
- add Data/j1j2_square_Coll_Ferromag.ccm (convergence checked, NR vs. DI)
- add Data/*.txt files (documentation, remarks , datas, known samples)
...
see ChangeLog in tgz-package
Documentation
The documentation is available in the documentation-path.
Also have an eye to the publication list.
Frequently asked questions (FAQ)
Q: What about performance?
A: Here are some results:
xxzSquare6-6 Oct04 (np=num_parallel_cpus, realtime,usertime,systime)
np=2 PM-600MHz 21s,6s,0s + 28m,14m,0s + 49s,14s,7s (old=slow)
+ 24s,10s,1s (new=fast)
np=4 PM-600MHz 20s,4s,0s + 16m, 4m,0s + 30s, 5s,0s
np=2 PM-600MHz 11s,4s,0s + 16m, 8m,0s + 22s, 9s,1s
np=1 PM-600MHz 4s,3s,0s + 16m,16m,0s + 19s,17s,1s
no_np PM-600MHz 3s,3s,0s + 16m,16m,0s + 18s,17s,1s
no_np PM-600MHz 3s,3s,0s + 16m,16m,0s + 82s,76s,5s (no_collect)
no_np PM-600MHz 3s,3s,0s + 12m06s noDEBUG + static_arrays
no_np PM-1200MHz 2s,2s,0s + 5m56s noDEBUG + static_arrays
Sep05: no_np Celeron 2GHz = 41s
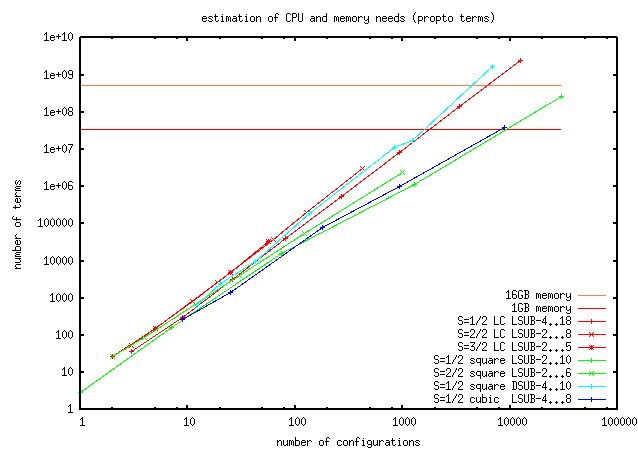 Q: Are there some test models to check correctness of the code?
A: Yes, you can find them in the ../Data path.
### model fundClusters bon1terms bond2terms D Eg M (Sep05)
xxzCubic-6-6 181 838 53620 D= -1.0 Eg= -1.80360459 M= 0.42859590 P2.6=1400s
kagome_j1j2-6 521 8163 458724 D= 0.0 Eg= -0.41827021 M= 0.27026674 P2.6=192s
S=1/2-LC-14 931 43537 4205e3 D= -1.0 Eg= -0.44255049 M= 0.14240498 ca.2h
S=1-chain-8-8 247 2466 56649 D= -1.0 Eg= -1.38346552 M= 0.53325176 P2.6=76s
Q: Are there some test models to check correctness of the code?
A: Yes, you can find them in the ../Data path.
### model fundClusters bon1terms bond2terms D Eg M (Sep05)
xxzCubic-6-6 181 838 53620 D= -1.0 Eg= -1.80360459 M= 0.42859590 P2.6=1400s
kagome_j1j2-6 521 8163 458724 D= 0.0 Eg= -0.41827021 M= 0.27026674 P2.6=192s
S=1/2-LC-14 931 43537 4205e3 D= -1.0 Eg= -0.44255049 M= 0.14240498 ca.2h
S=1-chain-8-8 247 2466 56649 D= -1.0 Eg= -1.38346552 M= 0.53325176 P2.6=76s
(potential) Bugs
- not known, please report (please try the latest version)
 Q: Are there some test models to check correctness of the code?
A: Yes, you can find them in the ../Data path.
### model fundClusters bon1terms bond2terms D Eg M (Sep05)
xxzCubic-6-6 181 838 53620 D= -1.0 Eg= -1.80360459 M= 0.42859590 P2.6=1400s
kagome_j1j2-6 521 8163 458724 D= 0.0 Eg= -0.41827021 M= 0.27026674 P2.6=192s
S=1/2-LC-14 931 43537 4205e3 D= -1.0 Eg= -0.44255049 M= 0.14240498 ca.2h
S=1-chain-8-8 247 2466 56649 D= -1.0 Eg= -1.38346552 M= 0.53325176 P2.6=76s
Q: Are there some test models to check correctness of the code?
A: Yes, you can find them in the ../Data path.
### model fundClusters bon1terms bond2terms D Eg M (Sep05)
xxzCubic-6-6 181 838 53620 D= -1.0 Eg= -1.80360459 M= 0.42859590 P2.6=1400s
kagome_j1j2-6 521 8163 458724 D= 0.0 Eg= -0.41827021 M= 0.27026674 P2.6=192s
S=1/2-LC-14 931 43537 4205e3 D= -1.0 Eg= -0.44255049 M= 0.14240498 ca.2h
S=1-chain-8-8 247 2466 56649 D= -1.0 Eg= -1.38346552 M= 0.53325176 P2.6=76s